Using next-generation sequencing (NGS) technologies for forensic investigations has gained interest over the past few years due to the possibility of obtaining more information from a DNA sample left at the crime scene, especially in the presence of mixed contributors. The development of new workflows to be used with the MiSeq® FGx Sequencing System opens up the possibility for forensic laboratories to perform multiple types of DNA analysis, including SNP-based kinship analysis, with a single instrument, avoiding the outsource of precious samples. Forensic laboratories can now perform STR analysis, mitochondrial control region and whole genome analysis, and now kinship analysis for forensic genetic genealogy (FGG), also known as investigative genetic genealogy, with the ForenSeq® Kintelligence kit from as little as 1 ng of DNA.
Written by: Joanna Antunes, Verogen
The ForenSeq Kintelligence kit analyzes 10,230 SNPs, and through the ForenSeq Universal Analysis Software (UAS), a report formatted for upload to GEDmatch PRO™ can be generated, making the process extremely streamlined.
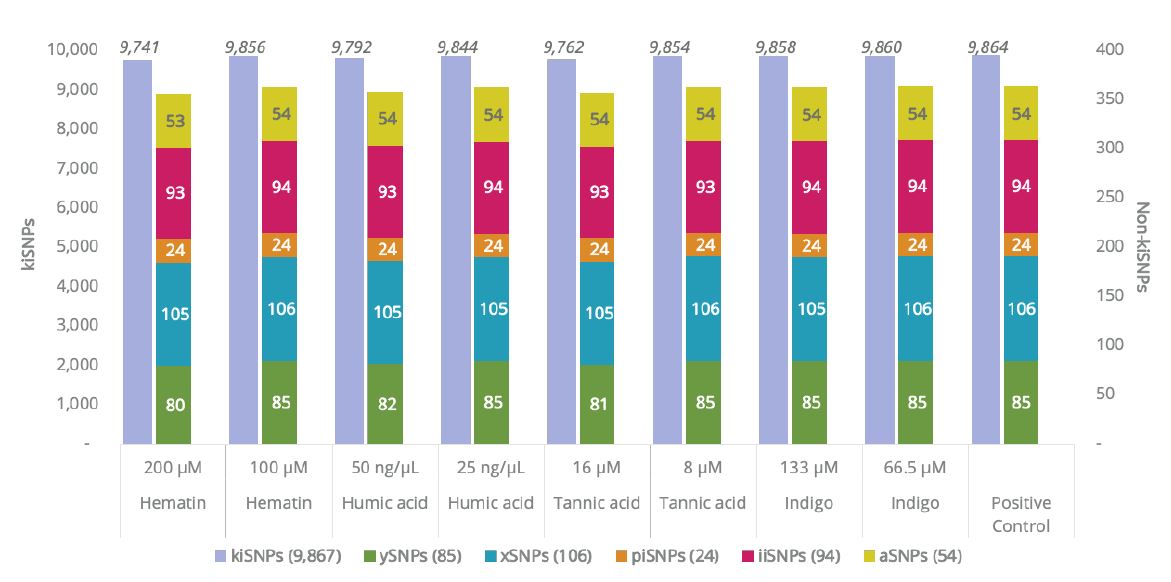
Forensic samples are notoriously challenging and at times it is impossible to obtain 1 ng of input DNA. Additionally, the majority of cases being investigated through FGG are cold cases or missing persons cases where degraded DNA samples are commonly present. At ISHI 32, we presented results obtained from testing degraded and low input DNA with ForenSeq Kintelligence. Additionally, we performed an inhibitors test, where commonly found inhibitors such as hematin, humic acid, tannic acid, and indigo were spiked into the PCR1 reaction to simulate situations where DNA extracted from blood, soil or plant material, and dyed clothes, contained co-extracted inhibitors.
The results (Figure 1) show that ForenSeq Kintelligence is extremely tolerant to the presence of inhibitors, with a SNP call rate of 99% even for the sample with higher loci dropouts (200 μM hematin).
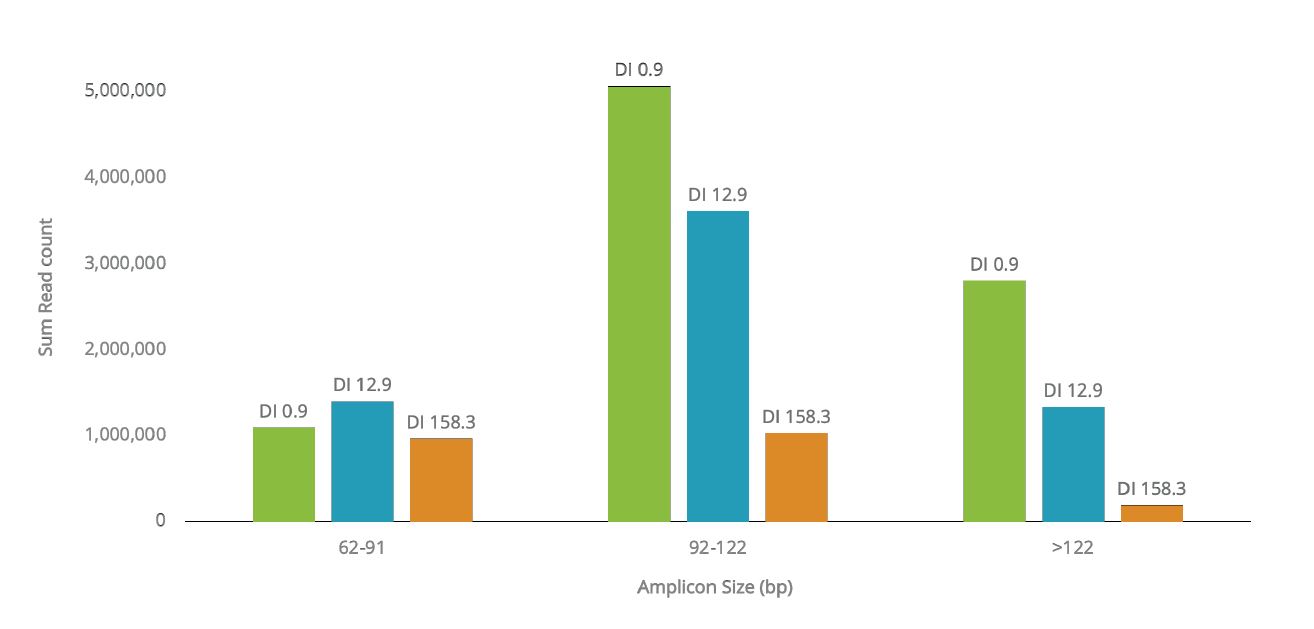
When degraded samples were tested, we observed that even for samples with a degradation index of 158.3 (as measured by the InnoQuant HY real-time qPCR kit), the majority of kiSNPs were still typed (9,728 SNPs out of the 9,867), making it possible for kinship analysis and upload to GEDmatch PRO. The majority of SNPs present in ForenSeq Kintelligence (5,257 SNPs) have amplicon lengths ranging from 92-122 bp, which likely allows the high call rate observed (98.5%) for degraded samples. We also observed that the read count for all amplicons decreased for the more degraded samples, especially for amplicons longer than 122 bp (Figure 2). This further supports the hypothesis that amplicon size must be minimized when designing kits for degraded forensic samples.
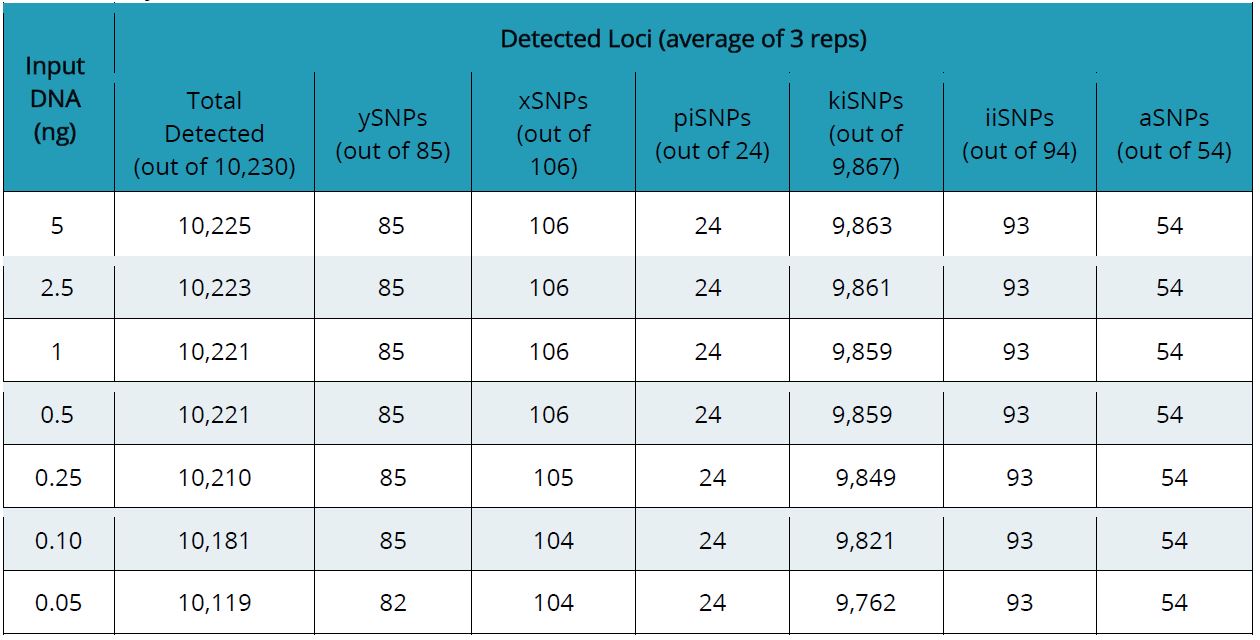
To determine the sensitivity of the ForenSeq Kintelligence kit, we ran a dilution series of the Positive Amplification Control (NA24385), testing DNA inputs from 5 ng to 0.050 ng into the PCR1 reaction. Although the recommended DNA input is 1 ng in 25 uL of sample, oftentimes crime scene samples contain little DNA, which can hinder the ability of complying with that input value.
The results (Table 1) show that even with only 50 pg of DNA input, the loci call rate is 99%, with kinship- informative SNPs demonstrating the highest number of locus dropouts (105 SNPs). No dropouts were observed for the phenotypic-informative SNPs or ancestry-informative SNPs. The high sensitivity of ForenSeq Kintelligence is possible due to the fact that the kit was designed with forensic samples in mind, making it a powerful tool for FGG cases.
The high number of loci typed for each sample (10,230) can feel overwhelming without a streamlined way to look at the data. The UAS is a software designed with ease-of-use in mind. For the ForenSeq Kintelligence kit, the user is able to obtain an overview of each sample from the main screen, with information regarding call rate, number of reads, biological sex and contributor status, as well as balance and how many SNPs were typed for each sub-category of SNPs (Figure 3).
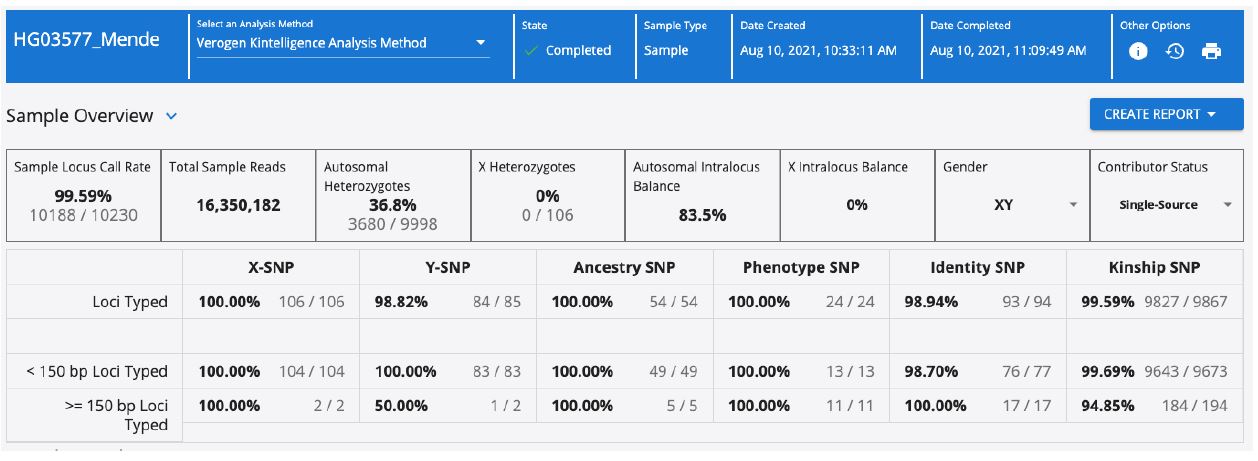
On UAS, the option of creating a report is intuitively displayed in a drop-down menu where one can choose “GEDmatch PRO”, “Sample”, or “Phenotype & Ancestry” reports. Multiple reports can be obtained for each sample, and the “GEDmatch PRO” report can be directly uploaded to GEDmatch. The “Sample” report is .xls format with the typed SNPs for each sub-category and the “Phenotype and Ancestry” report, showing the predictions for eye color, hair color, and biogeographical ancestry (Figure 4).
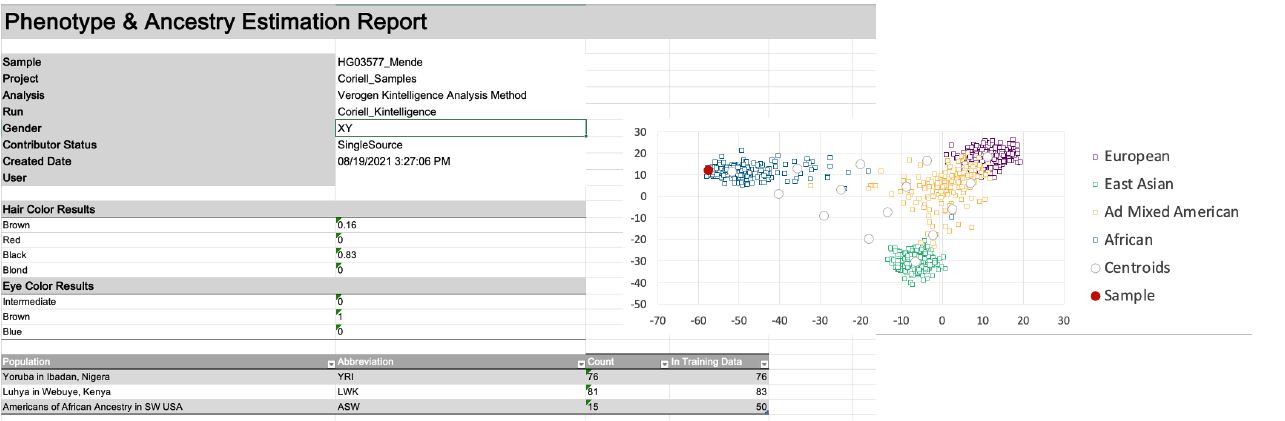
sample (1000 Genomes Mende sample) is displayed.
Overall, the ForenSeq Kintelligence kit displays good performance on inhibited, degraded, and low input DNA samples. The user-friendly UAS software simplifies data interpretation,
as well as the upload to databases such as GEDmatch PRO. As a full workflow, ForenSeq Kintelligence is a powerful tool that enables DNA analysts with the benefits of FGG without
needing to outsource their precious samples away from the lab.
Personally, I attended multiple talks at ISHI 32 and was moved by the stories of FGG providing emotional closure to families of missing persons and victims of violent crimes. I am humbled to know that my contribution to the development of the ForenSeq Kintelligence Kit can help the DNA analysts working on their cases. The developmental validation of the ForenSeq Kintelligence kit really highlights the performance of the kit, even in the challenging conditions that analysts encounter in their day-to-day work. I hope to continue to provide improved tools and more sensitive methods to facilitate their analysis and to allow the progress of justice.
WOULD YOU LIKE TO SEE MORE ARTICLES LIKE THIS? SUBSCRIBE TO THE ISHI BLOG BELOW!
SUBSCRIBE NOW!