Today’s blog is written by David Mittelman, PhD and Michael Vogen (Othram, Inc.) Reposted from The ISHI Report with permission.
The search for answers began in 1994 when a fisherman discovered a body in the lake. The Snohomish County Medical Examiner’s office determined the deceased to be between 25 and 35 years old and the victim of a violent homicide. There were multiple facial reconstructions for the victim, in part because ethnicity assessment was inconclusive. Each artistic rendition was shared with the public in hopes that someone would recognize the victim and come forward with information. After a quarter of a century, Lake Stickney John Doe’s true identity remained unknown. No match in CODIS, no established ethnicity, and certainly no leads on who was behind this heinous crime.
There are hundreds of thousands of cold cases, like the case of the Lake Stickney John Doe, in the United States and the vast majority of them will not be solved using traditional forensic DNA testing frameworks. One such framework is the Combined DNA Index System (CODIS), an FBI program that supports DNA databases and infrastructure for using these databases to search unknown DNAs against a catalog of known felons. While CODIS will remain a critical and irreplaceable component of forensic DNA testing for the foreseeable future, countless solvable cases remain unsolved due to the simple fact that it was designed – from the start, when DNA testing was in its infancy – to identify those individuals who had already been identified by other methods. While effective in tracking repeat crime by convicted criminals, CODIS is largely ineffective for unidentified remains or recently imported criminals, as many of them are victims and not criminals. CODIS will not reveal the identification of all criminals, either. Many perpetrators of crimes have yet to have be caught and even those that are caught (and even convicted) may purposely or accidently be missing from CODIS.
Incredible advances in genetic testing have transformed what forensic professionals can learn from DNA left at crime scenes and a growing number of seemingly “unsolvable” cases are now being closed. These newer DNA testing methods rely on “reading” tens to hundreds of thousands of DNA markers across the genome. There was early (albeit limited) success in collecting genome-wide markers using SNP microarrays. However, while SNP microarrays (sometimes called SNP chips) power recreational DNA testing methods for ancestry and genealogy, they are not nearly as effective for degraded, contaminated, or mixed DNAs – the bread and butter of forensic DNA testing. Moreover, arrays provide little indication that they are failing as they fail, so even if a result is returned, it is difficult to assess confidence in the DNA markers that are measured. Inaccurate data can have catastrophic effects on downstream genealogy efforts and prevent otherwise straightforward paths to human identification.
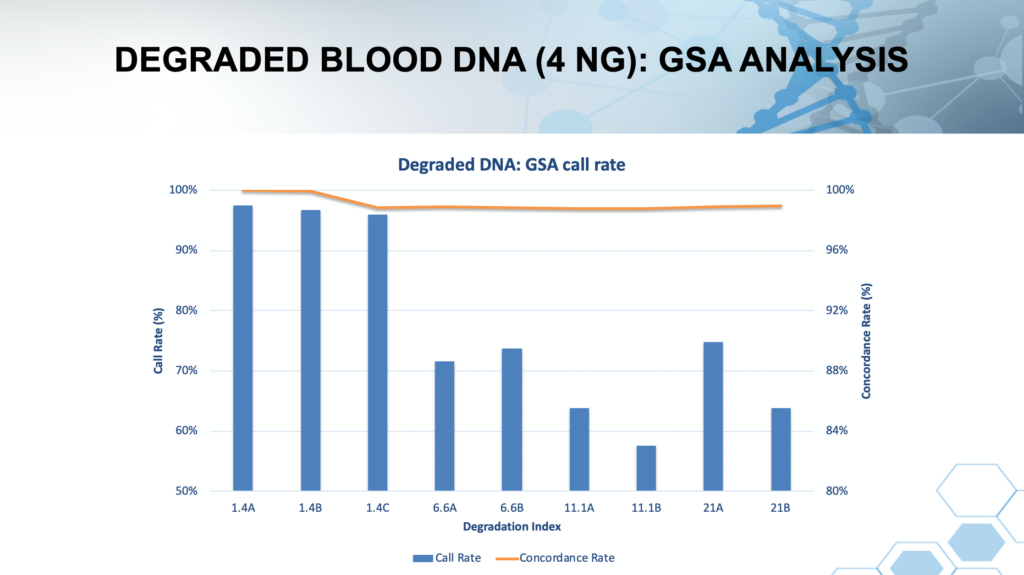
The plot in Figure 1 illustrates that even moderate DNA degradation has substantial impact on the performance of SNP chip call rates. In contrast, SNP call rates produced by Forensic-Grade Genome Sequencing® are insensitive to DNA degradation. Although not measured in the study summarized above, non-human contamination can have a substantial impact SNP chips as well. Bone extracts, for example, fail frequently on SNP chips, even at slight levels of degradation.
In addition to the above challenges, SNP chips require greater quantities of DNA than CODIS and other forensic methods. Coupling the increased quantity requirements with unpredictable failures for forensic evidence creates an alarming risk for fully consuming evidence without gaining the necessary information to establish a human identity. A key requirement for forensic DNA testing must be to extract the greatest number of informative markers while consuming the least amount of forensic evidence.
The better approach over SNP chips, especially for degraded and contaminated human remains, is to leverage massively parallel sequencing (MPS) techniques. Although more complex to utilize than SNP microarrays, MPS is more robust and accurate, and importantly, consumes less input DNA.

As an example, Othram’s Forensic-Grade Genome Sequencing® method is based on MPS. Our method combines MPS and advanced informatics to digitize genomes that otherwise fail with SNP chip and other non-sequencing methods. Genomic information is pieced together, leveraging multiple layers of quality data for every DNA position that is measured, to ensure accuracy.
A critical and under-appreciated advantage of massively parallel sequencing is the ability to detect novel genetic variations with exactly the same efficiency as previously seen variants. This can be used to minimize the types of error and bias intrinsic to microarrays and other methods, especially when working with imperfect forensic evidence. It is possible to combine sequence data (this cannot be done for SNP microarrays) so if insufficient data is generated from an initial test, results from multiple sequencing runs can be combined to produce an accurate representation of the DNA markers. The ability to iteratively and accurately collect DNA data from forensic evidence is critical for detecting distant genetic relationships and enabling human identification of an unknown person.
Case Study: Lake Stickney John Doe identified after 26 years
After nearly 26 years of working to identify the Lake Stickney John Doe, Snohomish County Sheriff’s Office Major Crimes Unit, Cold Case Team and the Snohomish County Medical Examiner’s Office positively identified the John Doe as Rodney Peter Johnson. Rodney was born in 1962 and his body was found in Lake Stickney in 1994. The Medical Examiner’s office had determined that Rodney had suffered a gunshot wound to his head.

For more than two decades, Rodney’s remains went unidentified. Ultimately, successful identification of Rodney was established using Forensic-Grade Genome Sequencing®. Detective Jim Scharf and Snohomish County Medical Examiner Lead Investigator, Jane Jorgensen worked with a lab to extract the victim’s DNA from a tooth, but the extracted DNA was heavily degraded and contaminated by non-human DNA. Less than 2% of the DNA was human. With just under a fifth of a nanogram of human DNA, the extract was sent to Othram for sequencing and genealogy.
Despite having about 20 cell’s worth of badly degraded human DNA to work with, Forensic-Grade Genome Sequencing®, paired with a combination of proprietary enrichment methods enabled the reconstruction of a genealogical profile for the Lake Stickney John Doe. Detective Scharf and Investigator Jorgensen used this genetic reconstruction and good old-fashioned detective work to identify Rodney Peter Johnson.
After uploading the genealogical profile to a public genealogical database, a match was identified. Usually, one match is not enough to point to a single person. However, this match had included another clue in their public database profile. They belonged to a distinctive direct maternal lineage. The high-resolution genealogical profile allowed investigators to perfectly match up this uncommon maternal lineage signature. The investigators then found a record for a missing person that appeared to match Mr. Johnson. Interestingly, although the missing person was reported in 1996, Rodney had last been seen in 1987. He was only 25 years old. It’s likely that after his murder, his body had been in the lake for as many as seven years before being discovered.
Investigators reached out to next-of-kin and used legacy DNA testing to confirm the identity established from the genealogical profile. With an identity now confirmed, investigators are now working to better understand what might have happened in the days leading up the Johnson’s death. The homicide investigation would have been intractable.
Concluding Thoughts
Incredible advances in forensic science—like the ability to identify suspects through a family tree or obtaining a full DNA profile from a single strand of rootless hair or touch DNA used to be science fiction. Today, however, the tools available to forensic professionals are unprecedented. Scientists continue to push the limits of what sorts of DNA evidence can be probed for identifying information. New methods that can sort out mixtures of human genomes or enable the enrichment of human genomes from non-human contaminating DNA. Caution must be exercised in consuming evidence. Fully consuming evidence using an inappropriate or inadequate can truly render a cold case unsolvable. As methods improve, we must invest in techniques that do not fully consume evidence and leave material for future testing and confirmations.
WOULD YOU LIKE TO SEE MORE ARTICLES LIKE THIS? SUBSCRIBE TO THE ISHI BLOG BELOW!
SUBSCRIBE NOW!


