Today’s blog is written by guest bloggers, François-Xavier Laurent, PhD, DNA Database Manager, INTERPOL and Susan Hitchin, PhD, DNA Unit Coordinator, INTERPOL. Reposted from The ISHI Report with permission.
The identification of human remains belonging to missing persons persists as a challenging process in forensic genetics. When unidentified human remains are discovered, a comparison between the post-mortem DNA profile (obtained from the remains), and the ante-mortem DNA profile (conventionally obtained from personal items of the missing person or from a previous medical sample) is the most reliable method for identification [1].
A number of countries have established national missing person DNA programmes, which are very effective when the disappearance of individuals and the discovery of unidentified human remains occur within the same country. However, many countries have unsolved missing person investigations and unidentified human remains that cannot be identified using their national systems alone. International cooperation in missing person investigations is therefore highly advised in light of increased global migration, the consequences of growing transnational crime and human trafficking, the vulnerability of migrants and refugees, and the latter’s high risk for being victims of a criminal act.
INTERPOL, the world’s largest international police organization, is mandated to assist in international investigations, including missing persons, by connecting its 194 member countries. The Organization works primarily with national law enforcement agencies via the INTERPOL National Central Bureau in each country but can also cooperate with other international entities involved in Disaster Victim Identification situations and missing person cases. INTERPOL provides the legal framework and technical infrastructure for a secure exchange of information and access to its 19 databases, thereby enabling police around the world to collaborate (Figure 1).

Created in 2002, the INTERPOL DNA database currently contains more than 250,000 profiles contributed by 86 member countries. National Central Bureaus and international entities can submit a DNA profile from offenders, crime scenes, missing persons and unidentified human remains, with an automated search result provided within minutes. There is no nominal data attached to the profile and member countries retain ownership of their information, in line with INTERPOL’s data processing rules. Countries can also choose with whom they wish to make their data available for comparison. The INTERPOL DNA database has enabled investigators around the world to link offenders to different types of crime including rape, murder and armed robbery as well as to formally identify missing persons found deceased outside their national borders from where they were reported missing.
However, in many cases, direct DNA matches are not possible as ante-mortem DNA profiles are either unavailable or insufficient to confirm the missing person’s identity. This could be due to the lack of available personal items or medical records, often key to missing person investigations, or due to the displacement of populations and the destruction or lack of property. Consequently, in the majority of cases, ante-mortem DNA data can only be obtained through relatives donating biological samples to the requesting authorities.
While most laboratories have the capacity and experience to perform relatively simple kinship testing, such as paternity tests, the evaluation of complex kinship scenarios is far more challenging [2]. Specialized computer software is often required to undertake comparisons of ante-mortem and post-mortem data to perform complex kinship calculations with large datasets of DNA profiles. This software computes likelihood ratios (LR), which are the optimal basis for statistical decisions, regardless of whether there is a hypothesis about prior probabilities [3]. Using specific allele frequencies from the reference population to which the missing person belongs, the fit of a missing person for any given pedigree (e.g. parent, child, or sibling of the missing person) is measured by comparing two hypotheses; H1 and H2. H1 supports that the individual is related to a defined pedigree, whereas H2 states that they are unrelated [2]. Although this method is relatively straightforward to implement in a national environment where the missing person and the unidentified human remains are reported within the same country, many challenges needed to be resolved before applying this method to an international configuration.
INTERPOL has addressed these challenges, and in June 2021, officially launched its 19th database, I-Familia, dedicated to international DNA kinship matching for missing persons (Figure 2).
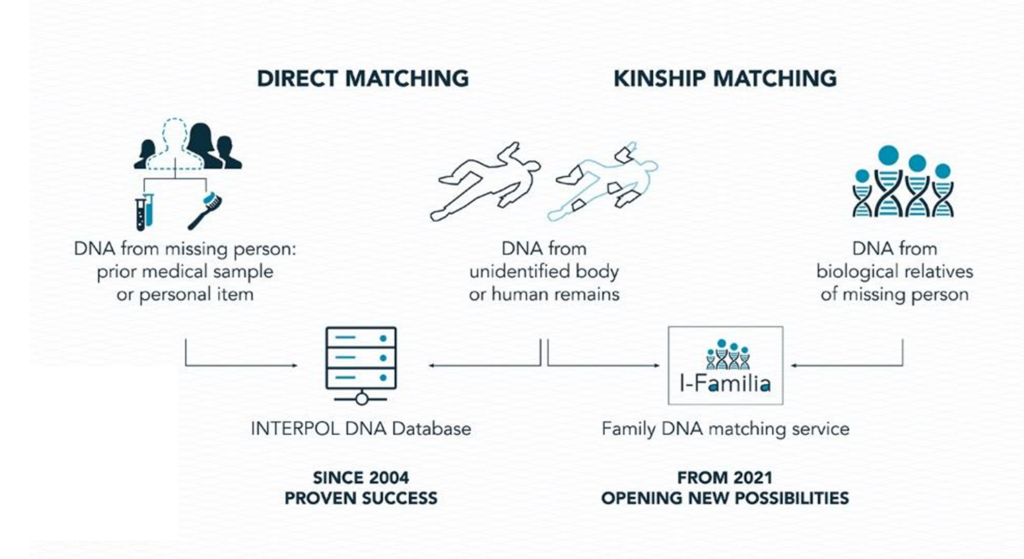
I-Familia, which stands for “INTERPOL Family Associated Matching to Identify Lost Individuals Abroad” is a pioneering service that is free-of-charge and available to all 194 member countries to help find potential biological relationships between DNA profiles from biological relatives of missing persons and unidentified human remains globally. I-Familia is composed of three components. First, a dedicated DNA database hosts anonymized DNA profiles from the biological relatives of missing persons and from the unidentified human remains. This is a stand-alone database with no links to other INTERPOL databases housing criminal data. Secondly, the state-of-the-art forensic DNA matching software BONAPARTE [4] is used to handle LR computations for any family pedigree with the available DNA data (autosomal STRs, Y-STRs, mitochondrial DNA) with all unidentified human remains DNA profiles. Thirdly, interpretation guidelines have been established to help users with the LR interpretation and the decision-making process that leads to the rejection or reporting of a potential biological relationship.
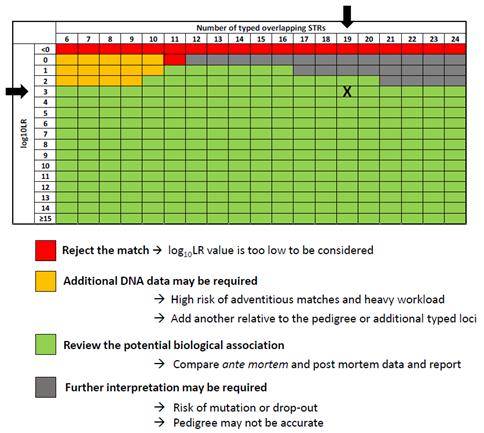
DNA kinship matching, based on autosomal STR profiles, requires the use of allele frequencies from the appropriate reference population. As information on the ancestry of DNA profiles is very often lacking or incorrectly reported, I-Familia enables LR computations using worldwide allele frequencies [5] and a theta correction factor to account for the degree of relatedness of alleles with a closely shared ancestry. To streamline the decision-making process and the review of potential biological relationships, extensive pedigree simulations were carried out to accurately determine optimal LR thresholds. Based on the 10 most frequently encountered scenarios in missing person investigations (depending on the availability of DNA profiles from biological relatives such as parents, children or siblings) and the number of overlapping STR loci (ranging between 6 and 24 loci), the LR thresholds help prevent the reporting of false-positive matches while minimizing the rejection of false-negative matches. Interpretation tables, specifically designed to reflect the expected number of adventitious matches for each type of match, are extremely useful to determine the best outcome (review the match, reject the match or require additional DNA information).
When the strength of the DNA evidence is sufficient, and ante-mortem and post-mortem data are compatible (if available), a potential biological relationship report is issued to both sources of data who can then cooperate on a bilateral basis to confirm the identification using their national procedures (Figure 3).
I-Familia has already been successful to help member countries with the identification of missing persons found in another country from where they were reported missing. The first confirmed match was found between the children’s DNA profiles of an Italian missing person and that of a body found in the Adriatic Sea by the Croatian police in 2004, therefore closing a case that had gone cold 16 years earlier.
By late 2021, over 12,000 active Yellow Notices – international police alerts for missing persons – had been issued by the INTERPOL General Secretariat highlighting the need for greater international cooperation. I-Familia is a humanitarian tool which, enabled by INTERPOL’s global reach, opens up vast new possibilities to identify missing persons and to provide families with answers.
With the purpose of providing scientific credibility and transparency to our member countries, the INTERPOL DNA Unit has recently published an article on the developmental validation of our interpretation guidelines for international DNA kinship matching [6]. This article describes the scientific rationale behind I-Familia and will allow forensic DNA experts around the world to replicate INTERPOL’s method and use this process nationally in the event of a missing person investigation for whom the genetic ancestry is unknown or uncertain.
For any additional information on I-Familia, please consult the I-Familia brochure [7] or contact the INTERPOL DNA unit at DNA@interpol.int.
References:
- Recommendations on the Use of DNA for the Identification of Missing Persons and Unidentified Human Remains by the INTERPOL DNA Monitoring Expert Group, (2017). http://www.interpol.int.
- M.D. Coble, J. Buckleton, J.M. Butler, T. Egeland, R. Fimmers, P. Gill, L. Gusmão, B. Guttman, M. Krawczak, N. Morling, W. Parson, N. Pinto, P.M. Schneider, S.T. Sherry, S. Willuweit, M. Prinz, DNA Commission of the International Society for Forensic Genetics: Recommendations on the validation of software programs performing biostatistical calculations for forensic genetics applications, Forensic Sci. Int. Genet. 25 (2016) 191–197. https://doi.org/10.1016/j.fsigen.2016.09.002.
- A. Collins, N.E. Morton, Likelihood ratios for DNA identification., Proc. Natl. Acad. Sci. 91 (1994) 6007–6011. https://doi.org/10.1073/pnas.91.13.6007.
- https://www.bonaparte-dvi.com/
- J. Buckleton, J. Curran, J. Goudet, D. Taylor, A. Thiery, B.S. Weir, Population-specific F values for forensic STR markers: A worldwide survey, Forensic Sci. Int. Genet. 23 (2016) 91–100. https://doi.org/10.1016/j.fsigen.2016.03.004.
- FX. Laurent, A. Fischer, R. Oldt, S. Kanthaswamy, J. Buckleton, S. Hitchin, Streamlining the decision-making process for international DNA kinship matching using worldwide allele frequencies and tailored cutoff log10LR thresholds, Forensic Sci. Int. Genet. 56 (2021). https://doi.org/10.1016/j.fsigen.2021.102634.
- https://www.interpol.int/How-we-work/Forensics/I-Familia
WOULD YOU LIKE TO SEE MORE ARTICLES LIKE THIS? SUBSCRIBE TO THE ISHI BLOG BELOW!
SUBSCRIBE NOW!


